Tutorial
This section provides a tutorial on how to use the AtomPacker package to pack nanoparticle atoms inside a target supramolecular cage. For detailed reference documentation of the functions and classes contained in the package, see the API reference.
In this example, we will use the ZOCXOH cage structure from the ZOCXOH.pdb file.
1. Load cage structure
First, import AtomPacker package on Python and create a AtomPacker.Cage object:
>>> import AtomPacker
>>> cage = AtomPacker.Cage()
The Cage object will be used to load the cage structure from the PDB file (eg., ZOCXOH.pdb):
>>> cage.load('ZOCXOH.pdb')
If you want to preview the cage structure, you can use the Cage.preview method.
>>> cage.preview()
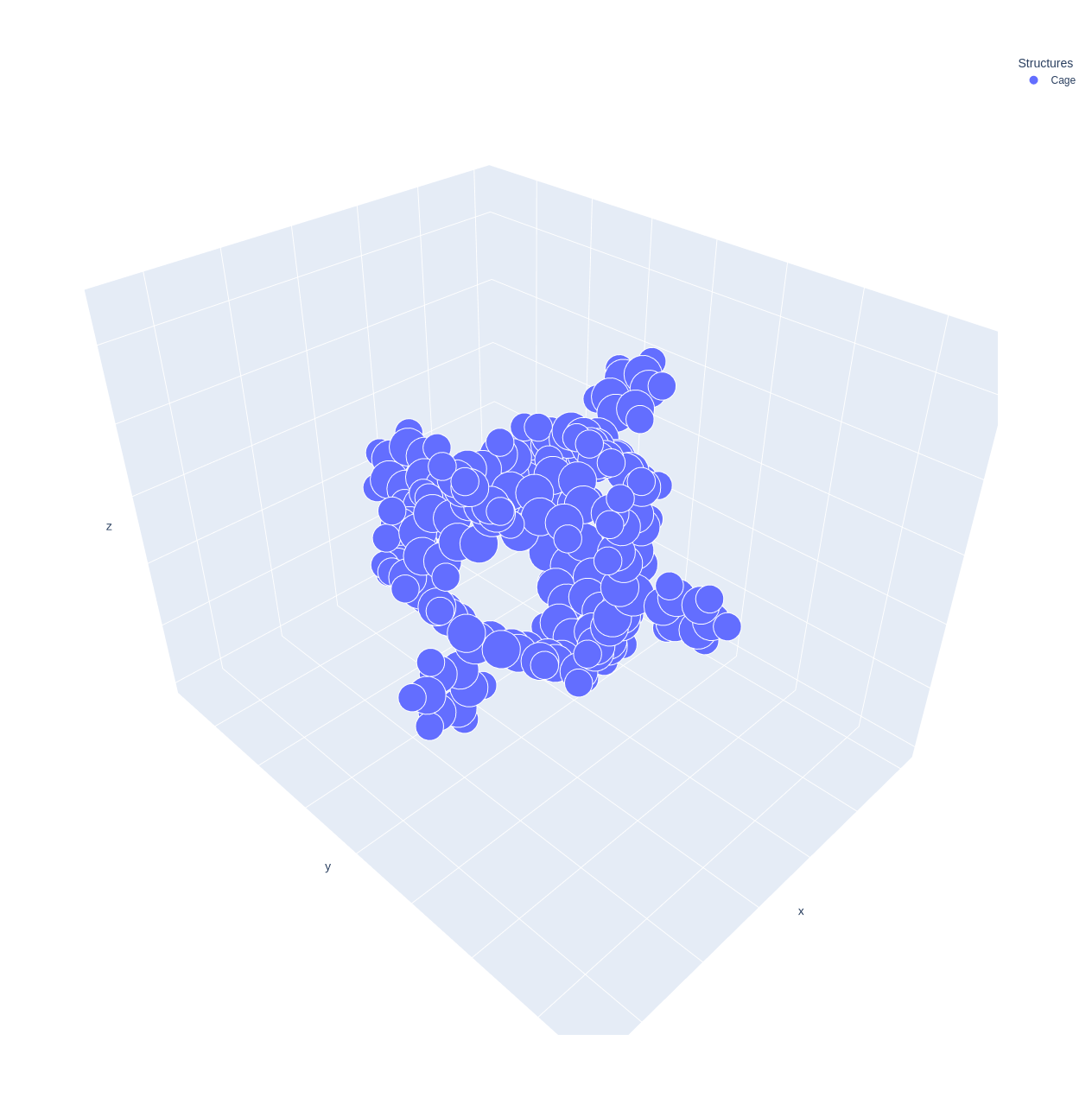
2. Detect cavity
Next, we will detect the cavity inside the cage structure. The Cage.detect_cavity method will detect the cavity using the pyKVFinder detection parameters (step, probe_in, probe_out, removal_distance, volume_cutoff, surface).
>>> cage.detect_cavity(step=0.6, probe_in=1.4, probe_out=10.0, removal_distance=1.0, volume_cutoff=5.0)
The detected cavity will be stored in the cavity attribute of the Cage object.
If you want to preview the cavity structure for quality control in your cavity detection, you can use the Cavity.preview method.
>>> cage.cavity.preview()
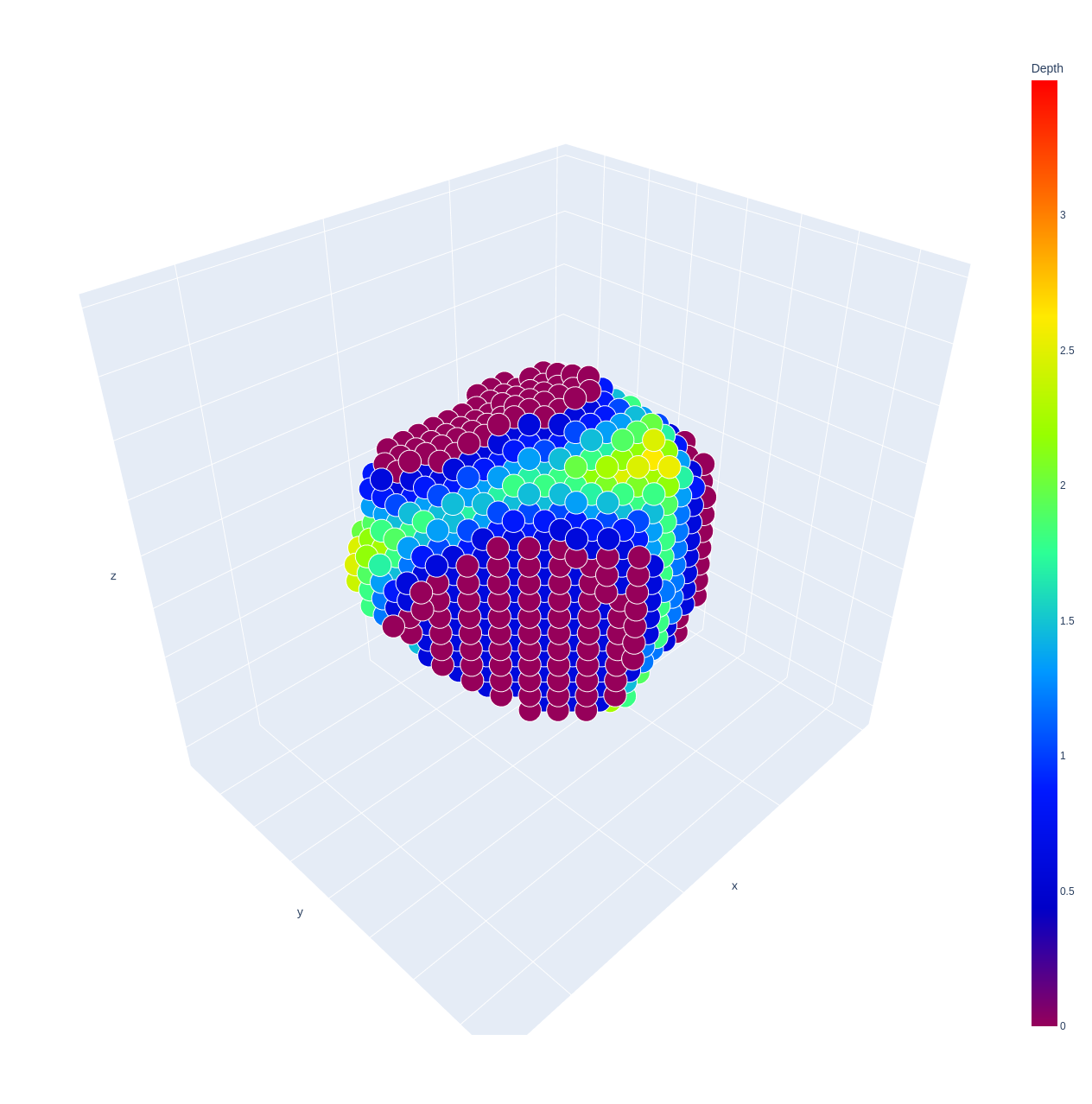
You can access the cavity coordinates using the coordinates attribute of the Cavity object.
>>> print(cage.cavity.coordinates)
[[-11.998 28.644 13.149 ]
[-11.998 28.644 13.749 ]
[-11.998 28.644 14.349 ]
...
[ -1.1980003 30.443998 14.349 ]
[ -1.1980003 30.443998 14.948999 ]
[ -0.59800035 29.844 13.749 ]]
You can access the cavity grid using the grid attribute of the Cavity object.
>>> print(cage.cavity.grid)
[[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
...
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]]
You can access the cavity volume using the volume attribute of the Cavity object.
>>> print(cage.cavity.volume)
531.58
You can also save the cavity structure using the Cavity.save method.
>>> cage.cavity.save("cavity.pdb")
3. Detect cavity openings
Next, we will detect the openings of the cavity. The Cage.Cavity.detect_openings method will detect the openings.
>>> cage.cavity.detect_openings()
The detected openings will be stored in the openings attribute of the Cavity object.
If you want to preview the openings structure, you can use the Openings.preview method.
>>> cage.cavity.openings.preview()
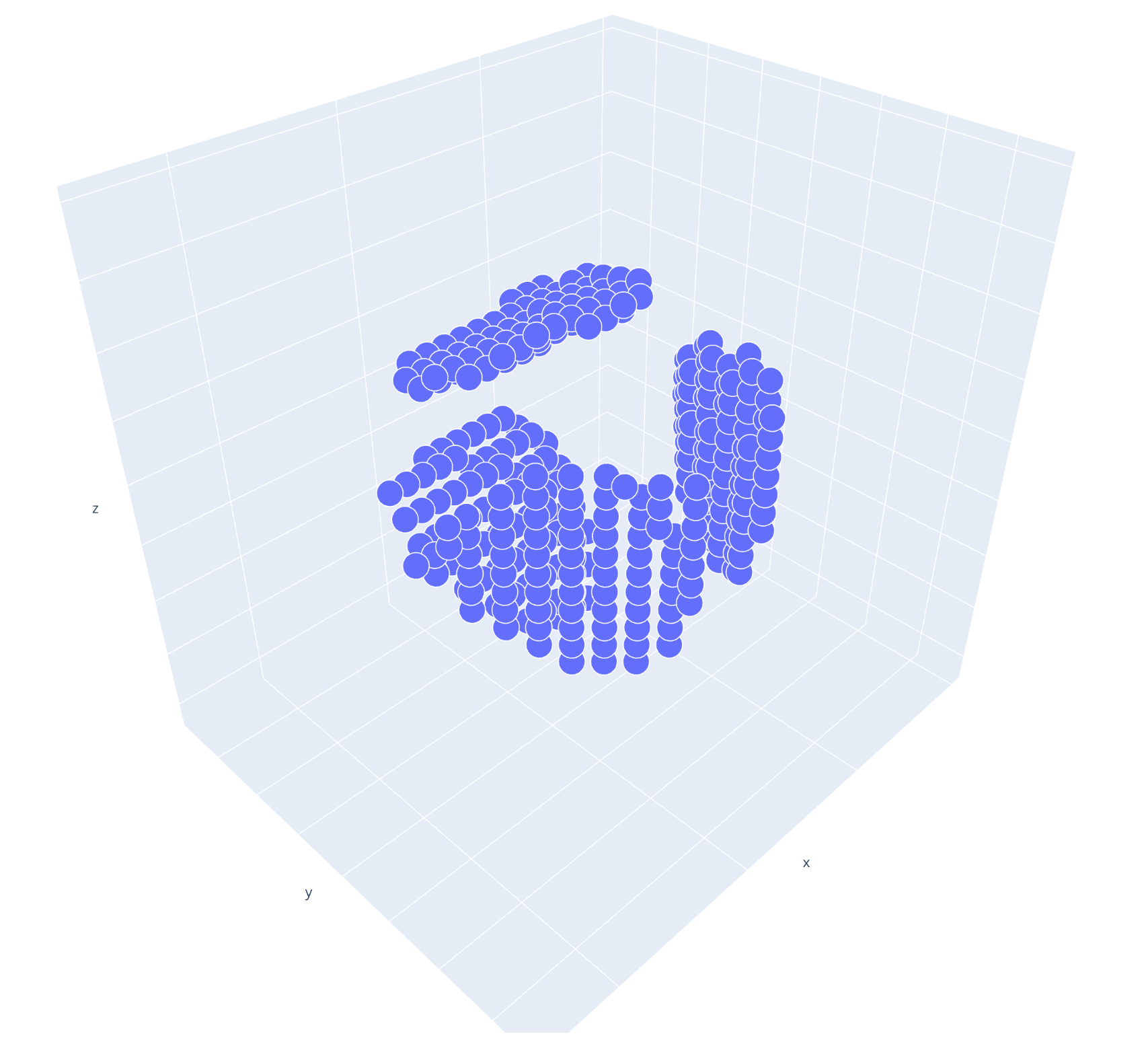
You can access the openings coordinates using the coordinates attribute of the Openings object.
>>> print(cage.cavity.openings.coordinates)
[[-11.998 28.644 13.149 ]
[-11.998 28.644 13.749 ]
[-11.998 28.644 14.349 ]
[-11.398001 29.244 12.549 ]
[-11.398001 29.244 13.149 ]
[-11.398001 29.244 13.749 ]
[-11.398001 29.244 14.349 ]
[-11.398001 29.244 14.948999 ]
[-11.398001 29.844 15.549 ]
[-10.798 29.844 11.948999 ]
[-10.798 29.844 12.549 ]
[-10.798 29.844 13.149 ]
[-10.798 29.844 13.749 ]
[-10.798 29.844 14.349 ]
[-10.798 29.844 14.948999 ]
[-10.798 30.443998 14.349 ]
[-10.798 30.443998 14.948999 ]
[-10.798 30.443998 15.549 ]
[-10.798 30.443998 16.149 ]
[-10.198 30.443998 11.349 ]
[-10.198 30.443998 11.948999 ]
[-10.198 30.443998 12.549 ]
[-10.198 30.443998 13.149 ]
[-10.198 30.443998 13.749 ]
[ -10.198 31.043999 10.749 ]
...
[ -1.1980003 29.844 13.749 ]
[ -1.1980003 30.443998 14.349 ]
[ -1.1980003 30.443998 14.948999 ]
[ -0.59800035 29.844 13.749 ]]
You can access the openings grid using the grid attribute of the Openings object.
>>> print(cage.cavity.openings.grid)
[[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
...
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]
[[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
...
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]
[-1 -1 -1 ... -1 -1 -1]]]
You can access the openings areas using the areas attribute of the Openings object.
>>> print(cage.cavity.openings.areas)
{'OAA': 43.53, 'OAB': 41.04, 'OAC': 41.4, 'OAD': 44.74}
You can access the openings diameters using the diameters attribute of the Openings object.
>>> print(cage.cavity.openings.diameters)
{'OAA': 7.444737563025417, 'OAB': 7.228675599024421, 'OAC': 7.260311091959885, 'OAD': 7.547498740076157}
You can also save the openings structure using the Openings.save method.
>>> cage.cavity.openings.save("openings.pdb")
4. Pack the nanocluster
Now that the cavity and its openings have been detected, you can pack nanoparticle atoms inside the cavity. Use the Cage.pack method to pack atoms with the desired parameters (atom_type, lattice_type, a, b, c). For example, to pack a gold (Au) nanoparticle using a face-centered cubic (fcc) lattice.
>>> cage.pack(atom_type="Au", lattice_type="fcc", a=None, b=None, c=None, angles=[0.0], translations=[0.0])
Note
To apply an optimization algorithm to the packed cluster, set angles and translations. Translations should be a list or numpy array of angles in degrees, and translations should be a list or numpy array of translation values in Angstroms. For example:
angles = [-75, -50, -25, 0, 25, 50, 75]
translations = [-0.2, 0.0, 0.2]
The packed cluster will be stored in the cluster attribute of the Cage object.
If you want to preview the cluster structure for quality control, you can use the Cluster.preview method.
>>> cage.cluster.preview()
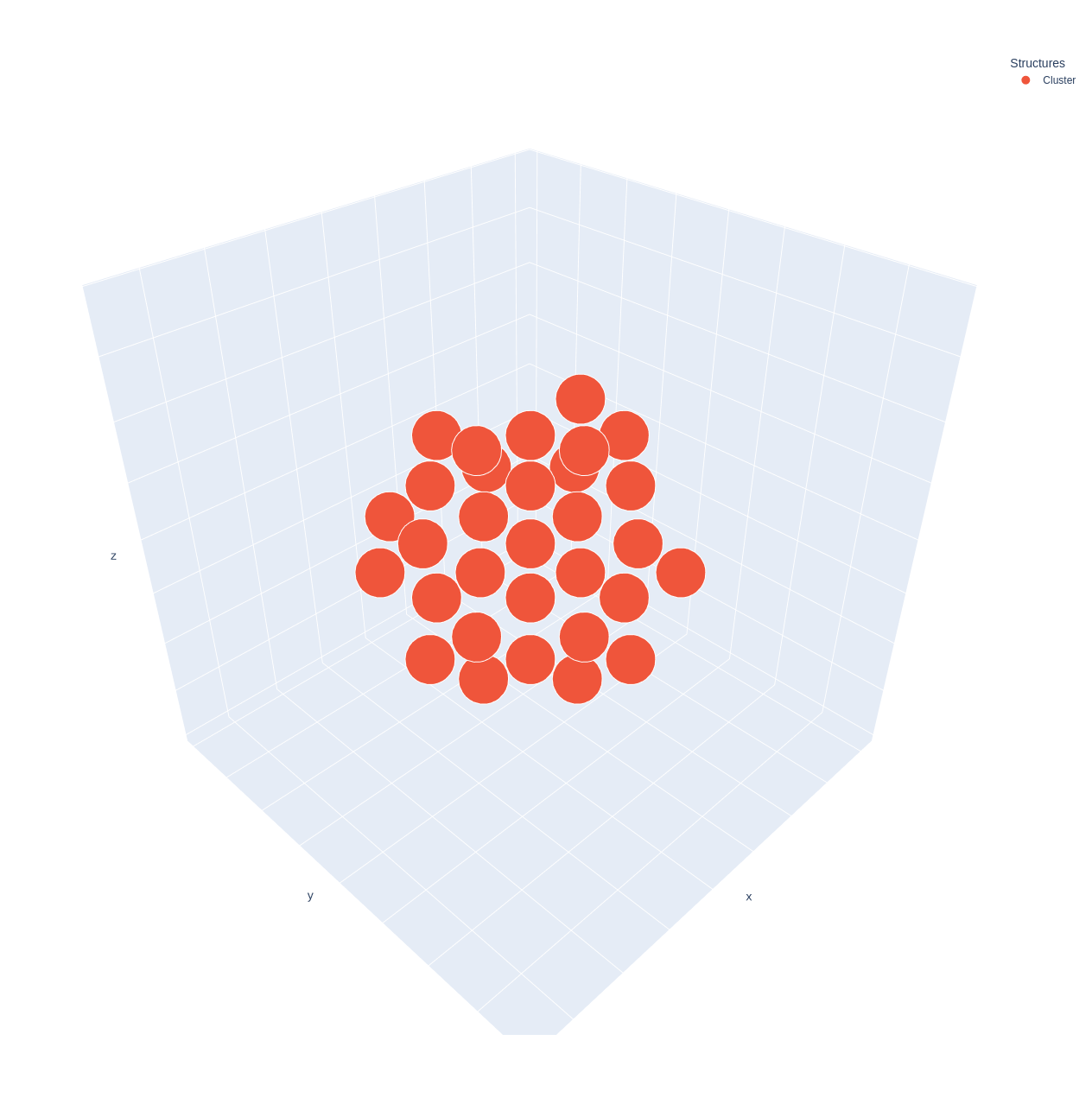
Also, you can preview the cage, cavity, openings and cluster structure using the Cage.preview method.
>>> cage.preview(show_cavity=True, show_cluster=True, show_openings=True)
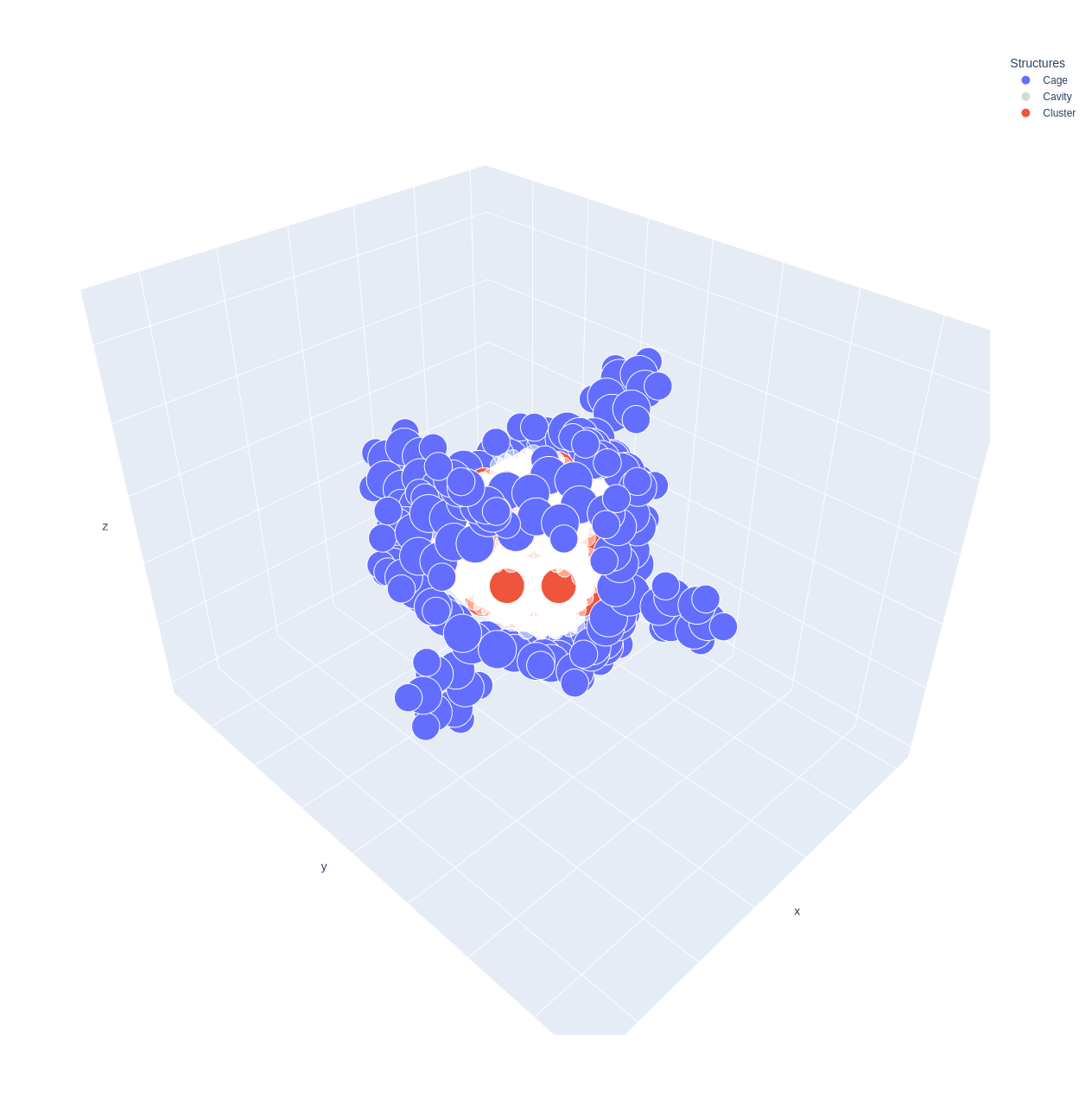
You can access the cluster coordinates using the coordinates attribute of the Cluster object.
>>> print(cage.cluster.coordinates)
[[ -8.56353 25.718132 13.873403]
[ -8.56353 27.758131 11.833403]
[-10.60353 27.758131 13.873403]
[-10.60353 29.798132 15.913403]
[ -8.56353 27.758131 15.913403]
[ -8.56353 29.798132 13.873403]
[ -8.56353 29.798132 17.953403]
[ -8.56353 31.838133 11.833403]
[ -8.56353 31.838133 15.913403]
[ -8.56353 33.87813 13.873403]
[ -6.52353 25.718132 15.913403]
[ -4.48353 25.718132 13.873403]
[ -6.52353 29.798132 11.833403]
[ -4.48353 27.758131 11.833403]
[ -4.48353 29.798132 9.793403]
[ -6.52353 27.758131 13.873403]
[ -6.52353 29.798132 15.913403]
[ -4.48353 27.758131 15.913403]
[ -4.48353 29.798132 13.873403]
[ -4.48353 29.798132 17.953403]
[ -6.52353 31.838133 9.793403]
[ -6.52353 33.87813 11.833403]
[ -4.48353 31.838133 11.833403]
[ -6.52353 31.838133 13.873403]
[ -6.52353 33.87813 15.913403]
[ -4.48353 31.838133 15.913403]
[ -4.48353 33.87813 13.873403]
[ -6.52353 31.838133 17.953403]
[ -2.44353 29.798132 11.833403]
[ -2.44353 27.758131 13.873403]
[ -2.44353 29.798132 15.913403]
[ -2.44353 31.838133 13.873403]]
You can also save the cavity structure using the Cluster.save method.
>>> cage.cluster.save("cluster.pdb")
Finally, you can access the summary of the cluster using the summary attribute of the Cluster object.
>>> print(cage.cluster.summary)
Au17
Atom Type Au
Atom Radius 1.441932
Cavity Volume (ų) 531.58
Diameter (maximum) 8.1568
Diameter (shape) 5.994511
Diameter (volume) 8.196339
Lattice Constants 4.0784
Lattice Type fcc
Maximum Number of Atoms 43
Number of Atoms 17
Also, you can access the optimization details using the log attribute of the Cluster object, using cage.cluster.log.