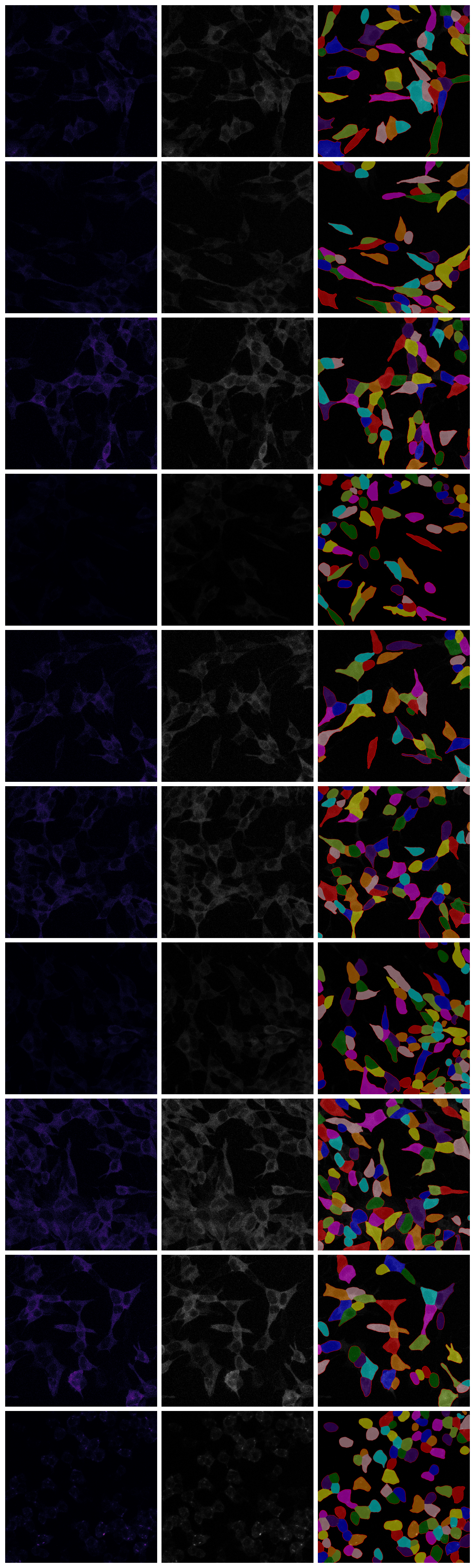
# Test cellpose to segment cells ```{python} from cellpose import modelsfrom glob import globimport imageio.v3 as iioimport skimage as skiimport numpy as npimport matplotlib.pyplot as plt``` ```{python} = models.CellposeModel(gpu= True , model_type= 'cyto3' )``` ```{python} = []= []= []for im_fn in glob('data/ome-tiff/*/*tif' )[:10 ]:= iio.imread(im_fn)# maximum intensity projection = im[0 ].max (axis= 0 )= im[1 ].max (axis= 0 )= im_cytoplasm# im_cyto_pre = ski.exposure.rescale_intensity(np.clip(im_cytoplasm,0,30), in_range=(0,30)) # im_cyto_pre = ski.exposure.rescale_intensity(np.clip(im_cytoplasm, 0, 20), out_range=(0.0,1.0)) = ski.filters.gaussian(im_cyto_pre, sigma= 0.5 )= ski.exposure.rescale_intensity(im_cyto_pre,out_range= (0 ,1 ))= im_nuclei= ski.filters.gaussian(im_nuclei_pre, sigma= 1 )= ski.exposure.rescale_intensity(im_nuclei_pre,out_range= (0 ,1 ))= 0 ))# break ``` ```{python} # fig, axs = plt.subplots(len(ims_cyto), 2, constrained_layout=True, figsize=(6, 3*len(ims_cyto))) # for idx in range(len(ims_cyto)): # axs[idx,0].imshow(ims_cyto[idx], cmap='gray') # axs[idx,1].imshow(ims_cyto_pre[idx], vmin=0, vmax=1, cmap='gray') # axs[idx,0].axis('off') # axs[idx,1].axis('off') ``` ```{python} = model.eval (ims_to_cellpose, channels= [0 ,1 ], diameter= 100 , normalize= True , min_size= 30 )``` ```{python} = plt.subplots(len (masks), 3 , constrained_layout= True , figsize= (9 , 3 * len (masks)))for idx, mask in enumerate (masks):0 ].imshow(ims_cyto[idx], cmap= 'magma' )1 ].imshow(ims_cyto_pre[idx], cmap= 'grey' )2 ].imshow(ims_cyto_pre[idx], cmap= 'grey' )2 ].imshow(ski.color.label2rgb(mask), alpha= 0.5 )2 ].contour(mask, linewidths= 0.1 , colors= 'red' )0 ].axis('off' )1 ].axis('off' )2 ].axis('off' )```