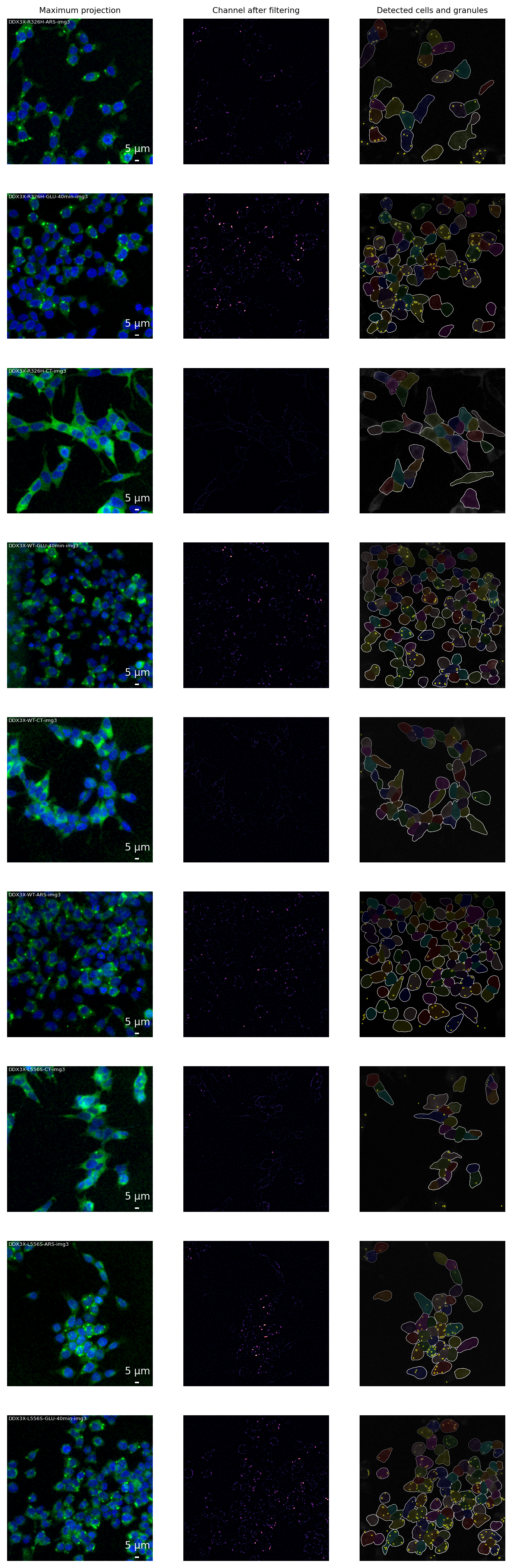
--- title: Image Analysis jupyter: python3 --- [ @stringerCellposeGeneralistAlgorithm2021 ] with the cyto3 model [ @stringerCellpose3OneclickImage2024 ] . Prior to segmentation, images were preprocessed with Gaussian smoothing (σ = 0.5 for GFP and σ = 1.0 for DAPI channels) and intensity rescaling to the [ 0,1 ] range. The model was configured with a target cell diameter of 100 pixels and minimum size threshold of 30 pixels. Cell masks were expanded by 5 pixels and border-touching cells were excluded from analysis.[ @waltScikitimageImageProcessing2014 ] . All measurements were converted to physical units using the microscope's calibrated pixel size (0.18 µm/pixel). For each cell, the number of contained granules was counted and normalized to cell area. The analysis pipeline was implemented in Python using cellpose, scikit-image, numpy, pandas and plotting libraries matplotlib, seaborn and microfilm v0.2.1.```{python} # image analysis import imageio.v3 as iiofrom cellpose import modelsfrom skimage.exposure import rescale_intensityfrom skimage.filters import gaussian, difference_of_gaussiansfrom skimage.morphology import remove_small_objectsfrom skimage.measure import label, regionprops, regionprops_tablefrom skimage.color import label2rgbfrom skimage.segmentation import clear_border, expand_labels# general import pandas as pdimport numpy as npimport refrom glob import globfrom os.path import basename# plot libs import matplotlib.pyplot as pltimport seaborn as snsfrom microfilm import microplot``` ```{python} # pixel size = 0.18um = 0.18 = ["data/ome-tiff/ddx3x_R326H/*.ome.tif" , "data/ome-tiff/ddx3x_wt/*.ome.tif" , "data/ome-tiff/ddx3x_L556S/*.ome.tif" ]``` ```{python} def save_sample_images(fig_title, im_nuclei, im_cytoplasm, im_cytoplasm_filtered, im_granules_labeled, cells_labeled):= plt.subplots(1 ,3 , constrained_layout= True )= gaussian(im_cytoplasm, 3 )= gaussian(im_nuclei, 1 )= np.quantile(im_nuclei_g, [0.01 , 0.999 ])= np.quantile(im_cyto_g, [0.2 , 0.999 ]) = [im_nuclei_g, im_cyto_g], = ["pure_blue" , "pure_green" ], = axs[0 ],= fig_title, = 5 ,= 'um' , = 0.18 , scalebar_size_in_units= 5 ,= 10 , scalebar_thickness= 0.01 ,= 'limits' ,= [[min_nuclei, max_nuclei], [min_cyto, max_cyto]])= [im_cytoplasm_filtered], = ["magma" ], = axs[1 ], = 'limits' ,= [[0 ,0.005 ]])2 ].imshow(im_cytoplasm, cmap= 'gray' )2 ].imshow(label2rgb(cells_labeled), alpha= 0.1 )# axs[2].imshow(label2rgb(im_granules_labeled), alpha=0.1) 2 ].contour(cells_labeled, colors= 'white' , linewidths= 0.1 )2 ].contour(im_granules_labeled, colors= 'yellow' , linewidths= 0.1 )2 ].axis("off" )# this line is specific to these data # fig_basename = re.search(r'\[(.*?)]', im_fn).group(1) # fig.suptitle(f'Image: {fig_basename}', fontsize=8) 0 ].set_title('Maximum projection' , fontsize= 6 )1 ].set_title('Channel after filtering' , fontsize= 6 )2 ].set_title('Detected cells and granules' , fontsize= 6 )f'figures/ { fig_title} .png' , bbox_inches = 'tight' , pad_inches = 0 , dpi= 600 )# fig.savefig(f'figures/{fig_basename}.pdf', bbox_inches = 'tight', pad_inches = 0, dpi=600) # fig.savefig(f'figures/{fig_basename}.svg', bbox_inches = 'tight', pad_inches = 0, dpi=600) ``` ```{python} def get_treatment_from_image_filename(im_fn):if "ARS" in im_fn:return 'ARS' elif 'GLU-40min' in im_fn:return 'GLU-40min' elif 'CT' in im_fn:return 'CT' else :raise (f"Fail in extract group from filename" , im_fn)``` ```{python} def get_image_number_from_image_filename(im_fn):= re.search(r'img(\d+)' , im_fn)= match.group(1 ) if match else 'unknown' return int (image_number)``` ```{python} def get_group_from_path(im_path):if 'ddx3x_wt' in im_path:return 'WT' elif 'ddx3x_R326H' in im_path:return 'R326H' elif 'ddx3x_L556S' in im_path:return 'L556S' ``` ```{python} = models.CellposeModel(gpu= True , model_type= 'cyto3' )# Lists to store data for both dataframes = []= []# plot = plt.subplots(9 ,3 , figsize= (9 ,28 ))= 3 = 0 for im_path in IMAGES_PATH:for im_fn in glob(im_path):if get_treatment_from_image_filename(im_fn) == 'GLU-20min' :# skip 20min GLU continue #read image = iio.imread(im_fn)# maximum intensity projection = im[0 ].max (axis= 0 ) # nuclei is the first channel = im[1 ].max (axis= 0 ) # cytoplasm is the second channel # Process cytoplasm and find granules = difference_of_gaussians(im_cytoplasm, 3 )= 0.0009 # we use a fixed global threshould because all images were collected at the same intensity = im_cytoplasm_filtered > threshold= remove_small_objects(im_granules, 15 )= clear_border(label(im_granules))# Get granule properties = regionprops_table(im_granules_labeled, = ['label' , 'area' , 'centroid' , 'eccentricity' , 'perimeter' ])# match = re.search(r'imagem(\d+)', im_fn) # image_number = match.group(1) if match else 'unknown' 'area' ] = granule_props['area' ] * PIXEL_SIZE * PIXEL_SIZE'perimeter' ] = granule_props['perimeter' ] * PIXEL_SIZE= get_image_number_from_image_filename(im_fn)'image_number' ] = image_number# granule_props['group'] = basename(im_fn).split(".lif")[0] = get_treatment_from_image_filename(im_fn)'treatment' ] = image_treatment= get_group_from_path(im_path)'group' ]= image_group# Segment cells with cellpose # preprocessing to improve cellpose segmentation = gaussian(im_cytoplasm, sigma= 0.5 )= rescale_intensity(im_cyto_pre,out_range= (0 ,1 ))= gaussian(im_nuclei, sigma= 1 )= rescale_intensity(im_nuclei_pre,out_range= (0 ,1 ))= np.stack([im_cyto_pre, im_nuclei_pre], axis= 0 )= model.eval (im_to_cellpose, channels= [0 ,1 ], diameter= 100 , min_size= 30 )= clear_border(expand_labels(masks, 5 ))# Process each cell for cell in regionprops(cells_labeled):= cells_labeled == cell.label= im_granules_labeled[cell_mask]= len (np.unique(granules_in_cell[granules_in_cell > 0 ]))= cell.area * PIXEL_SIZE * PIXEL_SIZE'treatment' : image_treatment,'group' : image_group,'image_number' : image_number,'cell_label' : cell.label,'cell_area' : cell_area,'granule_count' : granule_count,'granules_per_area' : granule_count / cell_area if cell_area > 0 else 0 = f"DDX3X- { image_group} - { image_treatment} -img { image_number} " if image_number == example_number:# plot one image from each group = gaussian(im_cytoplasm, 3 )= gaussian(im_nuclei, 1 )= np.quantile(im_nuclei_g, [0.01 , 0.999 ])= np.quantile(im_cyto_g, [0.2 , 0.999 ]) = [im_nuclei_g, im_cyto_g], = ["pure_blue" , "pure_green" ], = axs[idx, 0 ],= fig_title, = 5 ,= 'um' , = 0.18 , scalebar_size_in_units= 5 ,= 10 , scalebar_thickness= 0.01 ,= 'limits' ,= [[min_nuclei, max_nuclei], [min_cyto, max_cyto]]) = [im_cytoplasm_filtered], = ["magma" ], = axs[idx,1 ], = 'limits' ,= [[0 ,0.005 ]])2 ].imshow(im_cytoplasm, cmap= "gray" )2 ].imshow(label2rgb(cells_labeled), alpha= 0.1 )2 ].contour(cells_labeled, colors= 'white' , linewidths= 0.1 )2 ].contour(im_granules_labeled, colors= 'yellow' , linewidths= 0.1 )2 ].axis("off" )# names and labels if idx == 0 :0 ].set_title('Maximum projection' , fontsize= 8 )1 ].set_title('Channel after filtering' , fontsize= 8 )2 ].set_title('Detected cells and granules' , fontsize= 8 )+= 1 # save all images to verify the quality # Create final dataframes = pd.concat(granule_data, ignore_index= True )= pd.DataFrame(cell_data)# Save figure with examples 'figures/examples.png' , bbox_inches = 'tight' , pad_inches = 0 , dpi= 600 )'figures/example.pdf' , bbox_inches = 'tight' , pad_inches = 0 , dpi= 600 )'figures/example.svg' , bbox_inches = 'tight' , pad_inches = 0 , dpi= 600 )``` ```{python} "output/granules_per_cell.csv" , index= False )"output/granule_area.csv" , index= False )```