# Plots of results
```{python}
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
```
```{python}
cells_df = pd.read_csv("output/granules_per_cell.csv")
granules_df = pd.read_csv("output/granule_area.csv")
```
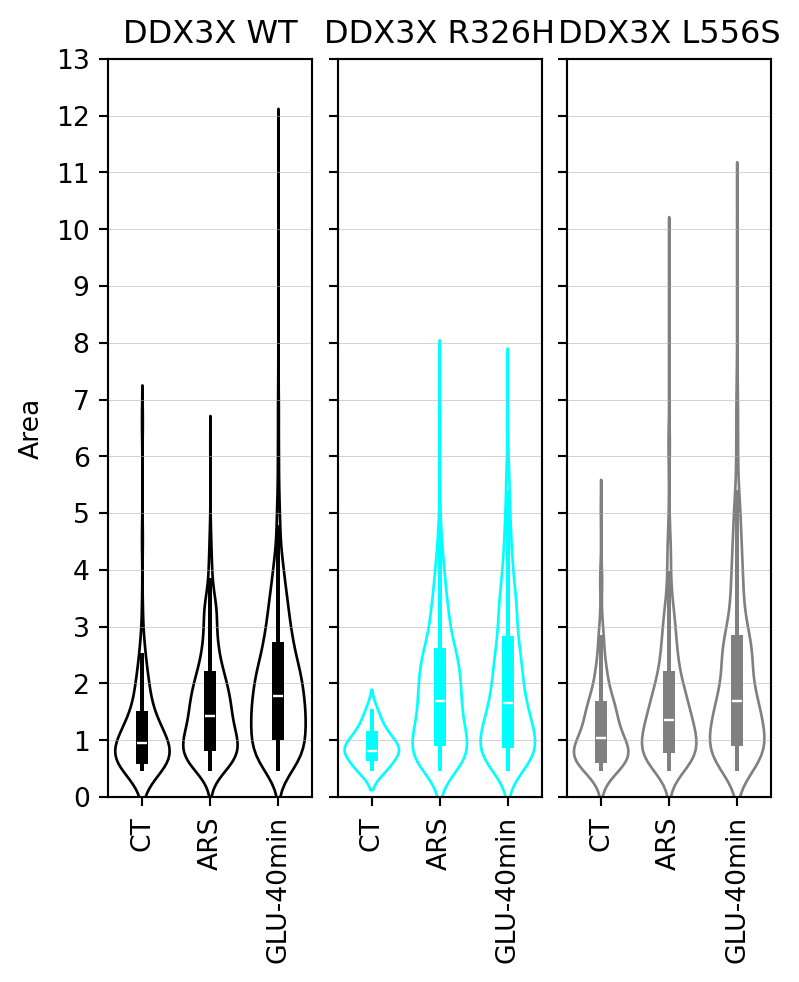
## Stress granule area
In this section, we present the results of the statistical analysis of stress granule area.
```{python}
#|fig-cap: "Violin plots of stress granule area."
fig, axs = plt.subplots(1,3, figsize=(4,5), sharey=True, constrained_layout=True)
axs[0].set_title('DDX3X WT')
axs[1].set_title('DDX3X R326H')
axs[2].set_title('DDX3X L556S')
xlabels = ['CT', 'ARS', 'GLU-40min']
sns.violinplot(data=granules_df[granules_df.group == 'WT'],
x='treatment', y='area',
fill=False,
inner='box',
ax=axs[0],
order=xlabels,
color='k',
density_norm='width',
linewidth=1)
sns.violinplot(data=granules_df[granules_df.group == 'R326H'],
x='treatment', y='area',
fill=False,
inner='box',
ax=axs[1],
order=xlabels,
color='cyan',
density_norm='width',
linewidth=1)
sns.violinplot(data=granules_df[granules_df.group == 'L556S'],
x='treatment', y='area',
fill=False,
inner='box',
ax=axs[2],
order=xlabels,
color='grey',
density_norm='width',
linewidth=1)
for ax in axs:
ax.set_xlabel('')
ax.set_ylabel('Area')
ax.set_yticks(range(0,14))
ax.set_ylim(0,13)
ax.set_xticklabels(xlabels, rotation=90)
ax.grid(axis='y', linewidth=.2)
plt.savefig("plots/stress_granules_area.png")
plt.savefig("plots/stress_granules_area.svg")
```
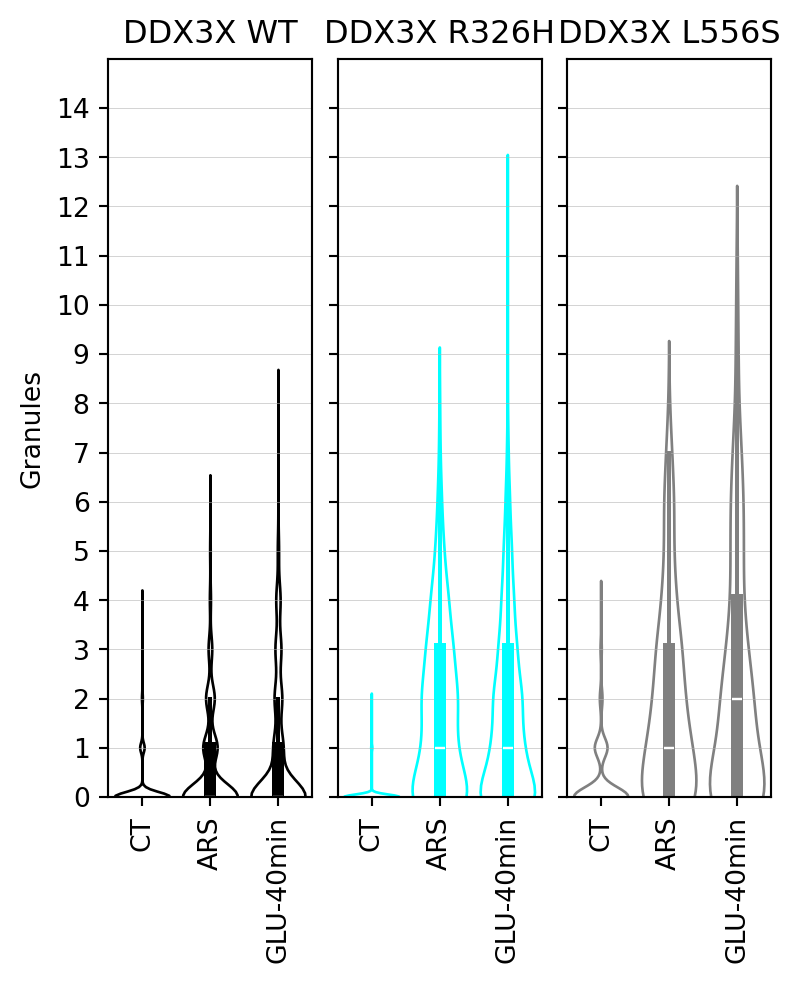
## Stress granule count per cell
In this section, we present the results of the statistical analysis of stress granule count per cell.
```{python}
#|fig-cap: "Violin plots of stress granule count per cell."
fig, axs = plt.subplots(1,3, figsize=(4,5), sharey=True, constrained_layout=True)
axs[0].set_title('DDX3X WT')
axs[1].set_title('DDX3X R326H')
axs[2].set_title('DDX3X L556S')
xlabels = ['CT', 'ARS', 'GLU-40min']
sns.violinplot(data=cells_df[cells_df.group == 'WT'],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[0],
order=xlabels,
color='k',
density_norm='width',
linewidth=1)
sns.violinplot(data=cells_df[cells_df.group == 'R326H'],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[1],
order=xlabels,
color='cyan',
density_norm='width',
linewidth=1)
sns.violinplot(data=cells_df[cells_df.group == 'L556S'],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[2],
order=xlabels,
color='grey',
density_norm='width',
linewidth=1)
for ax in axs:
ax.set_xlabel('')
ax.set_ylabel('Granules')
ax.set_yticks(range(0,15))
ax.set_ylim(0,15)
ax.set_xticklabels(xlabels, rotation=90)
ax.grid(axis='y', linewidth=.2)
plt.savefig("plots/stress_granules_per_cell.png")
plt.savefig("plots/stress_granules_per_cell.svg")
```
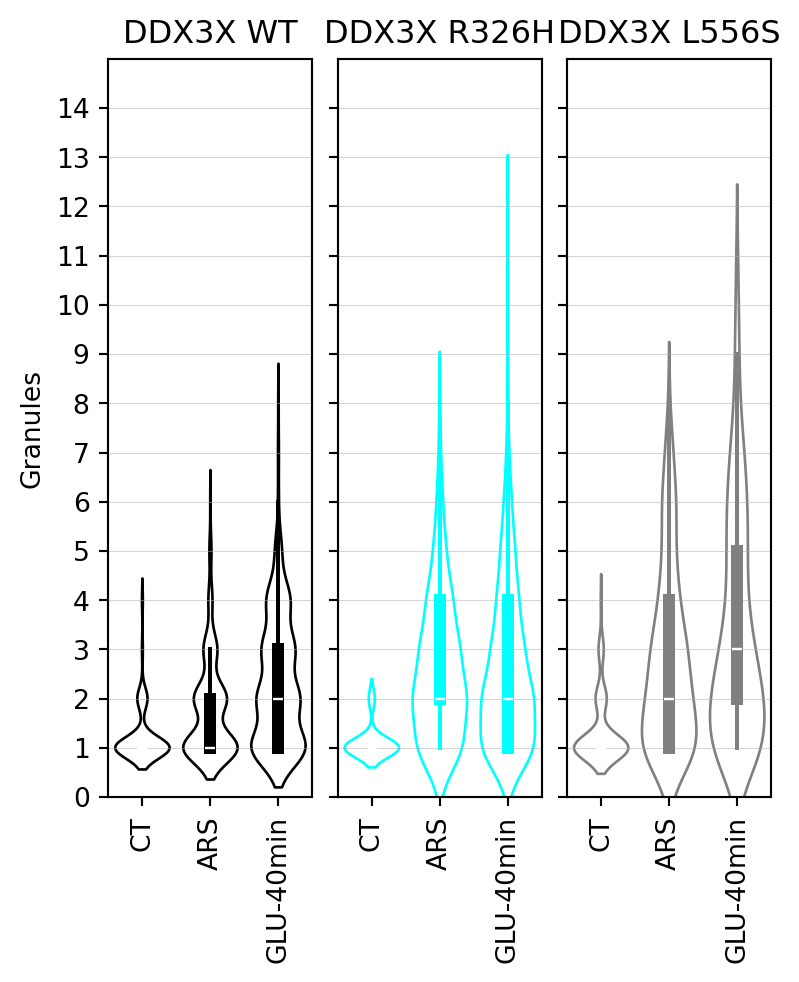
## Stress granule count per cell excluding cells without stress granules
In this section, we present the results of the statistical analysis of stress granule count per cell excluding cells with zero stress granules.
```{python}
#|fig-cap: "Violin plots of stress granule count per cell excluding cells with zero stress granules."
fig, axs = plt.subplots(1,3, figsize=(4,5), sharey=True, constrained_layout=True)
axs[0].set_title('DDX3X WT')
axs[1].set_title('DDX3X R326H')
axs[2].set_title('DDX3X L556S')
xlabels = ['CT', 'ARS', 'GLU-40min']
sns.violinplot(data=cells_df[(cells_df.group == 'WT')&(cells_df.granule_count > 0)],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[0],
order=xlabels,
color='k',
density_norm='width',
linewidth=1)
sns.violinplot(data=cells_df[(cells_df.group == 'R326H')&(cells_df.granule_count > 0)],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[1],
order=xlabels,
color='cyan',
density_norm='width',
linewidth=1)
sns.violinplot(data=cells_df[(cells_df.group == 'L556S')&(cells_df.granule_count > 0)],
x='treatment', y='granule_count',
fill=False,
inner='box',
ax=axs[2],
order=xlabels,
color='grey',
density_norm='width',
linewidth=1)
for ax in axs:
ax.set_xlabel('')
ax.set_ylabel('Granules')
ax.set_yticks(range(0,15))
ax.set_ylim(0,15)
ax.set_xticklabels(xlabels, rotation=90)
ax.grid(axis='y', linewidth=.2)
plt.savefig("plots/stress_granules_per_cell_wout_zeros.png")
plt.savefig("plots/stress_granules_per_cell_wout_zeros.svg")
```
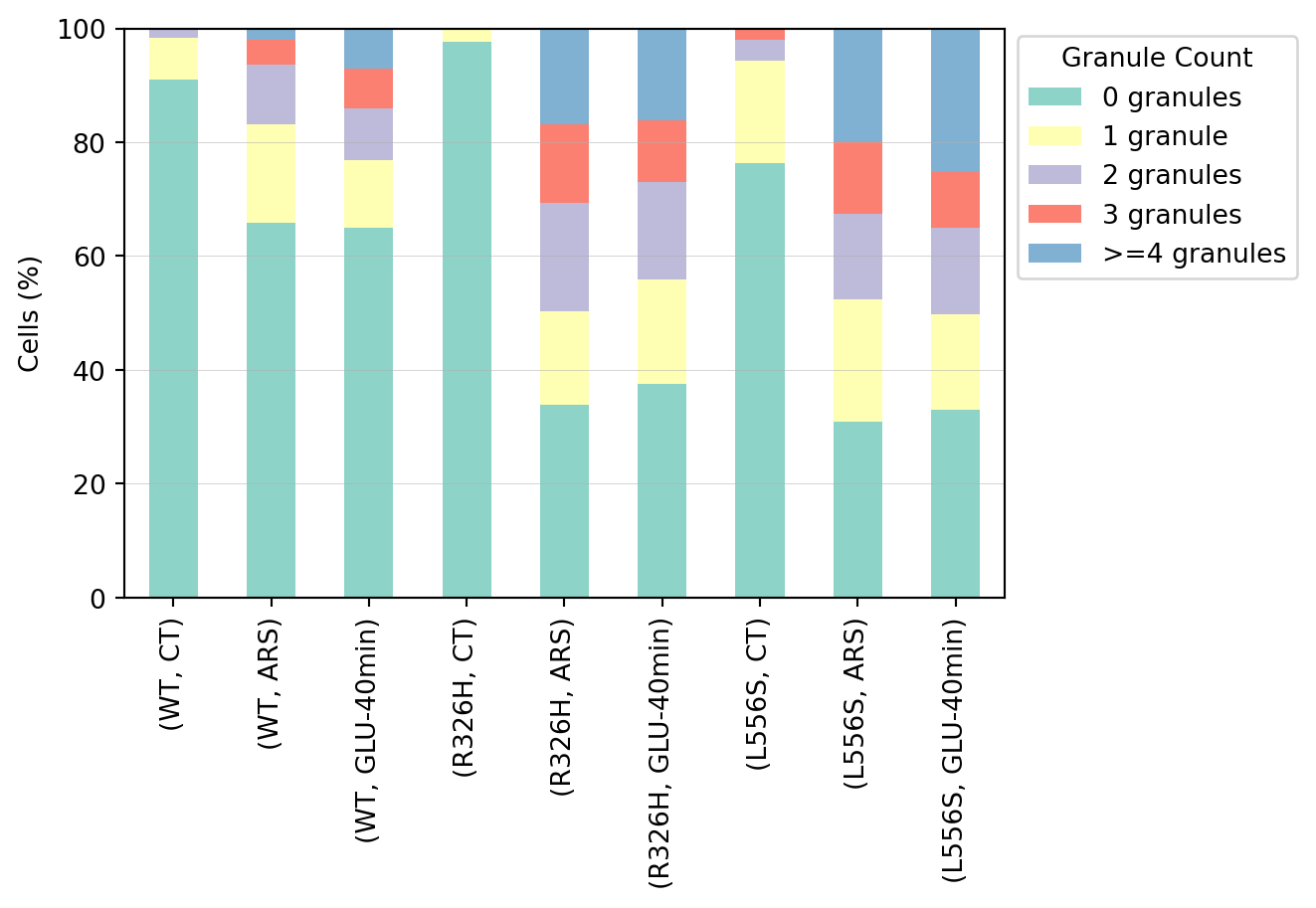
## Distribution of granule counts
In this section, we present the distribution of granules per cell.
```{python}
# Create categories for different granule counts
sns.set_palette('Set3')
def categorize_granules(count):
if count == 0:
return '0 granules'
elif count == 1:
return '1 granule'
elif count == 2:
return '2 granules'
elif count == 3:
return '3 granules'
else:
return ">=4 granules"
# Add category column
cells_df['granule_category'] = cells_df['granule_count'].apply(categorize_granules)
# Calculate percentages for each group and category
category_percentages = cells_df.groupby(['group', 'treatment', 'granule_category']).size().unstack()
category_percentages = category_percentages.div(category_percentages.sum(axis=1), axis=0) * 100
# category_percentages = category_percentages.sort_values('group', key=lambda x: pd.Categorical(x, categories=['CT', 'ARS', 'GLU-20min', 'GLU-40min'], ordered=True))
# Create stacked bar plotplt.grid(axis='y', linewidth=.2)2, 6))
category_percentages = category_percentages.sort_values('treatment', key=lambda x: pd.Categorical(x, categories=['CT', 'ARS', 'GLU-20min', 'GLU-40min'], ordered=True)).sort_values('group', ascending=False)
category_percentages.plot(kind='bar', stacked=True)
sns.violinplot(data=granules_df[granules_df.group == 'R326H'],
x='treatment', y='area',
fill=False,
inner='box',
ax=axs[1],
order=xlabels,
color='cyan',
density_norm='width',
linewidth=1)
# plt.xticks(rotation=45, ha='right')
plt.ylabel('Cells (%)')
# plt.xlabel('Group')
plt.legend(title='Granule Count', bbox_to_anchor=(1.0, 1.01), loc='upper left')
plt.tight_layout()
plt.xlabel('')
plt.ylim(0,100)
plt.grid(axis='y', linewidth=.2)
plt.savefig("plots/cells_with_stress_granules.png")
plt.savefig("plots/cells_with_stress_granules.svg")
```